Simulating Brownian Motion on a Computer.
The simulations used in Lab Books are all based on a simple core program that stores spatial information for each displayed particle and updates its location for each unit of time (called a time slice).
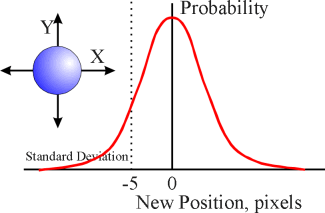 To move each particle, two random numbers are drawn: one is used to update the x-axis position, the other to update the y-axis position. The random numbers themselves are produced from an algorithm that calculates its output according to a bell-shaped (Gaussian) curve. The peak probability of this Gaussian distribution is set at zero, with decreasing probability of obtaining either a positive or negative value further and further from zero. For the simulations used here, the Gaussian distribution for single particles, lipids and water molecules has a standard deviation (SD) of 5 units, while the Gaussian distribution for the movement of larger ion/water assemblies and proteins is much more tightly clustered around the mean, having an SD of 2 units. As with all Gaussian distributions, +/- 1 SD accounts for about two-thirds of all the values; +/- 3 SD includes 99% of all possible values.
To move each particle, two random numbers are drawn: one is used to update the x-axis position, the other to update the y-axis position. The random numbers themselves are produced from an algorithm that calculates its output according to a bell-shaped (Gaussian) curve. The peak probability of this Gaussian distribution is set at zero, with decreasing probability of obtaining either a positive or negative value further and further from zero. For the simulations used here, the Gaussian distribution for single particles, lipids and water molecules has a standard deviation (SD) of 5 units, while the Gaussian distribution for the movement of larger ion/water assemblies and proteins is much more tightly clustered around the mean, having an SD of 2 units. As with all Gaussian distributions, +/- 1 SD accounts for about two-thirds of all the values; +/- 3 SD includes 99% of all possible values.
This random walk scheme used in Lab Book 2 approximates Brownian (diffusion) movement actually observed in a planar fluid medium. Each molecule in a real fluid, however, is vibrating and continuously colliding and interacting with its neighbors, but these events occur on a time scale faster than we can observe and simulate. During a period equivalent to our time slice, the "net" movement simulated is the average of a large number of such collisions and interactions. The average of many such random events is well described by the bell-shaped Gaussian curve used for our calculations. We also depicted lipids diffusing in the plane of a membrane about 2.5 more rapidly than integral proteins, which clearly underestimates their relative difference in velocity. Longer term interactions in our FRAP simulations, such as exist in lipid plaques or between IMP and their lipid "girdles", have been ignored, but we have simulated the tethering of certain IMP to peripheral membrane proteins of the cytoskeleton, albeit in a static manner. These simulations are misleading in one other way: because we have drawn the protein images much larger than their virtual locations and have shaded them appropriately, they occasionally seem to overlap and cluster in the third dimension; this clustered appearance is a simulation artifact.
The field of view in the FRAP experiments is considered a "window", and any particle leaving the window is replaced by another entering at a random location around the frame; all bleached particles diffusing out of the window are replaced by unbleached ones. (In this manner, our FRAP experiments simulate what an observer would see within a square "microscope field-of-view.") Otherwise, the only force "driving" the various movements are the Gaussian random numbers in the x and y directions. Using this approach, our simulations reproduce well the actual movement of lipid and IMP in a planar membrane.
The programs themselves are written in the language Java. They make use of the concept of "object-oriented" programming by making each element in the simulation responsible for its own properties. Thus, each particle knows how to draw itself, stores its position and charge, and updates its position based on the constraints placed on its motion.
 To move each particle, two random numbers are drawn: one is used to update the x-axis position, the other to update the y-axis position. The random numbers themselves are produced from an algorithm that calculates its output according to a bell-shaped (Gaussian) curve. The peak probability of this Gaussian distribution is set at zero, with decreasing probability of obtaining either a positive or negative value further and further from zero. For the simulations used here, the Gaussian distribution for single particles, lipids and water molecules has a standard deviation (SD) of 5 units, while the Gaussian distribution for the movement of larger ion/water assemblies and proteins is much more tightly clustered around the mean, having an SD of 2 units. As with all Gaussian distributions, +/- 1 SD accounts for about two-thirds of all the values; +/- 3 SD includes 99% of all possible values.
To move each particle, two random numbers are drawn: one is used to update the x-axis position, the other to update the y-axis position. The random numbers themselves are produced from an algorithm that calculates its output according to a bell-shaped (Gaussian) curve. The peak probability of this Gaussian distribution is set at zero, with decreasing probability of obtaining either a positive or negative value further and further from zero. For the simulations used here, the Gaussian distribution for single particles, lipids and water molecules has a standard deviation (SD) of 5 units, while the Gaussian distribution for the movement of larger ion/water assemblies and proteins is much more tightly clustered around the mean, having an SD of 2 units. As with all Gaussian distributions, +/- 1 SD accounts for about two-thirds of all the values; +/- 3 SD includes 99% of all possible values.