Studying
Cell Biology
Preface |
Introduction to Problem Solving | Problem
Sets | Acknowledgments
|
E.4.b.
The Temperature Shift Experiment |
Print
PDF
|
Next Part>>
| E.4.a. |
| E.4.b. |
| E.4.c. |
|
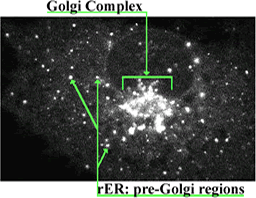
The movement of newly synthesized membrane protein through the complex array of intracellular membranes presents interesting questions of both sorting and motility. Recently, it has been possible to tag newly synthesized protein “naturally” with a fluorescent tag by inserting the nucleotide sequence for Green Fluorescent Protein (GFP, a jelly fish protein) to one end of the gene coding for the protein of interest. Following translation, GFP spontaneously folds into a fluorescent “tag” that in many instances does not inhibit the subsequent processing or ultimate structure or function of its chimeric partner. The figures and videos that follow were produced from time-lapse observations of the progression of newly synthesized, GFP-labeled vesicular stomatitis virus membrane protein(GFP-VSVP) from ER to the Golgi complex in cultured COS cells. Very thin optical sections of each cell was obtained by confocal microscopy. In these images, a faintly fluorescing ER network surrounds a collection of much brighter, large vesicles that constitutes the Golgi complex. In all instances the Golgi complex, when present, is situated adjacent to the nucleus. Images in the first video were captured by a digital camera about every 9 sec; they are displayed in the quick-time movie at a rate of about 9 frames a second and thus are speeded up about 81-fold. The second video lasts 18 sec, and runs at about 12 frames per sec; its images were captured about every 4 sec, and the movie therefore speeds up the natural process about 48-fold. Recall what you know about the steps of post-translational processing, click on the still image below to bring up the movie, watch the movement of GFP-VSVP into the Golgi several times (at least once, one frame at a time) and answer the questions in the right-hand frame. [You can learn more about these phenomena and the following experiments by consulting the video essay by Presley, et al. 1998 Mol Biol. Cell 9:1617-1626. PMID9658158] |
||
|
Questions A. Once again, describe the movement of the tagged protein as the temperature is raised. B. How do your observations compare with those made from watching the first video? C. How might you determine whether GFP-VSVP is diffusing randomly from ER to Golgi? D. If you think the movement is non-random, how might it directed. Turn the page to consider the results of pretreating the COS cells with nocodazole. |
|